HOW TO SEARCH and QUERY RuphiBase
The database entries correspond to putative Ruditapes trascripts, chracterised by bioinformatic analyses.
Individual transcripts are associated to different kinds of sequence and annotation by similarity information.
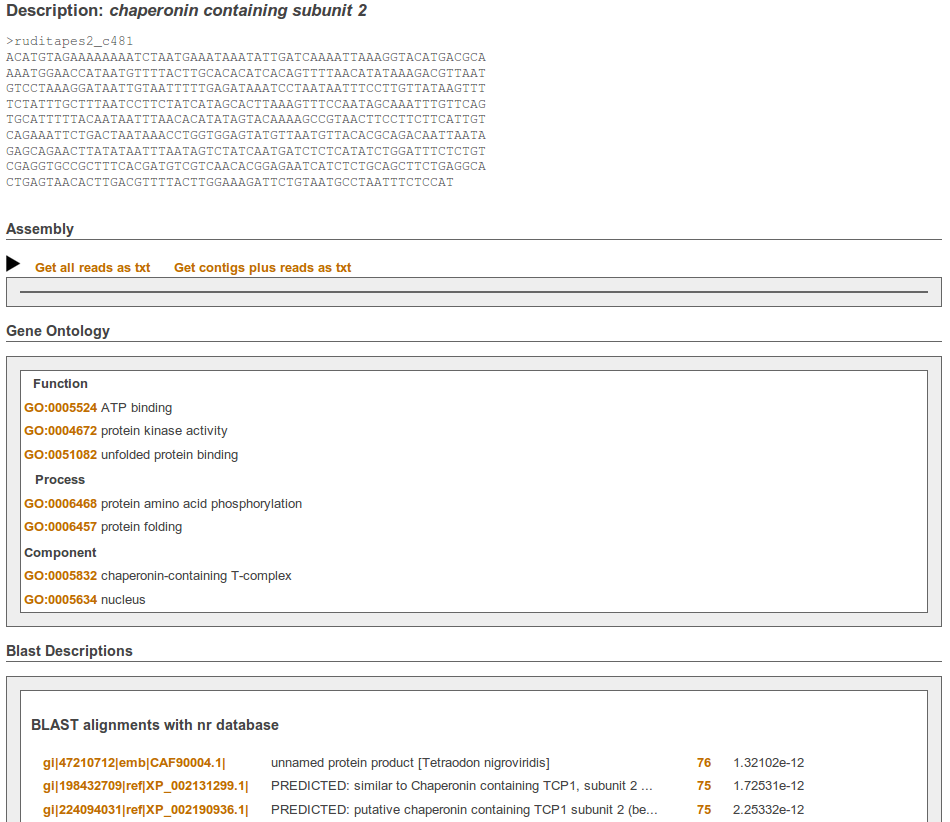
See a sample RuphiBase entry
http://compgen.bio.unipd.it/ruphibase/contig/ruditapes2_c481/
HOW TO SEARCH RuphiBase
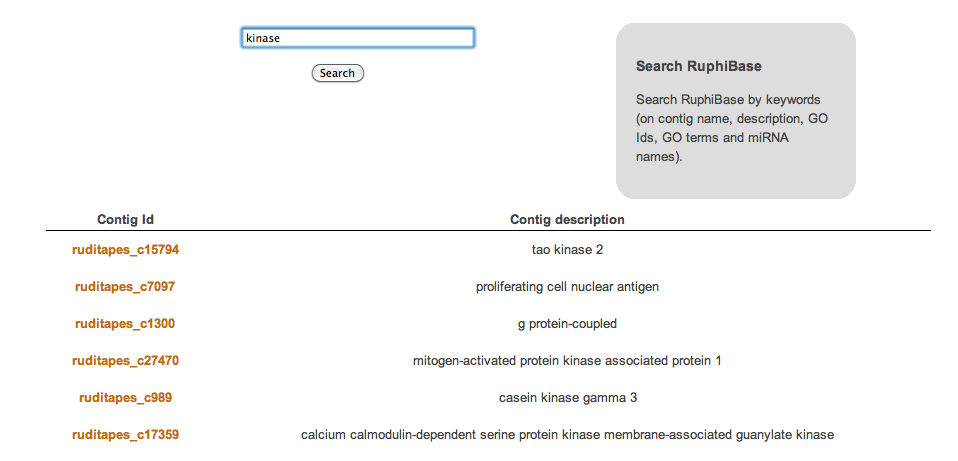
The result of such a search is a table listing accession numbers and descriptions of matching entries, linked to contig pages.
Screenshots of search form, results table and linked entry:

HOW TO BROWSE RuphiBase for MASSIVE DATA RETRIEVAL
Indeed, the the user can customise the retrieved information.
Simple query:
- using a given contig accession number
- retrieving contig description and sequence information
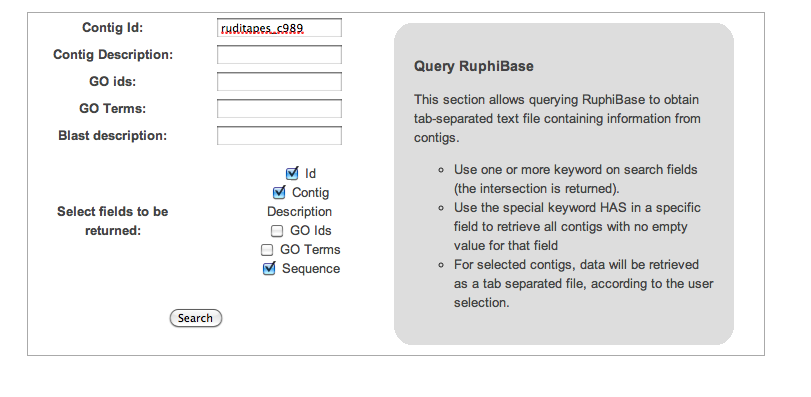
ruditapes_c989 casein kinase gamma 3 GTCGGGAAACCGGCCGGCCATTTGGTGTTTGGGTAACCCCGGGGA...
Advanced query:
- Using “growth” as keywords on description field
- using HAS on GO descriptions, to consider all the entries corresponding to contigs associated to GO terms
- etrieving both sequence and annotation information
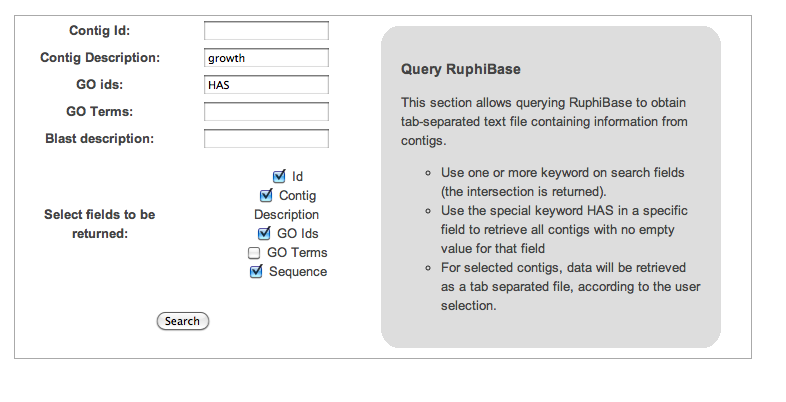
ruditapes_c28116 growth arrest-specific 8 GO:0008285,GO:0005929,GO:0005794,GO:0009434,GO:0005515,GO:0030317,GO:0005874 GGAAGACGCATGGAGAACAAATGGCTGANATCTCAGCCAAAAATAAAATGCTGGCTGAACCACTACAGAAAGCCCAGGAGGAG... ruditapes2_c2387 fibroblast growth factor receptor 2 GO:0044425,GO:0004713,GO:0006468,GO:0005886,GO:0005515,GO:0048522,GO:0050793,GO:0048513,GO:0007243,GO:0048869,GO:0004872 AAGGAATGGAATATCTTGCTGGAAGAGGGAT... ... ...
