CirComPara: A multi-method comparative bioinformatics pipeline to detect and study circRNAs from RNA-seq data
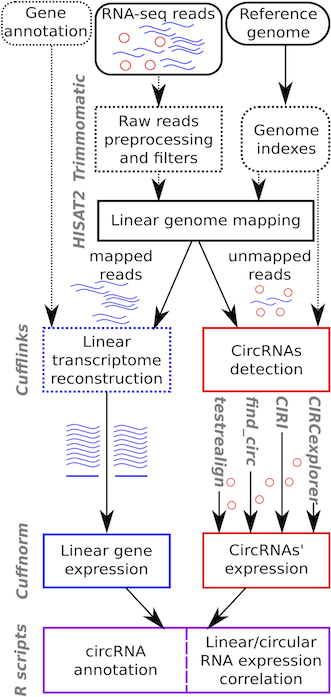
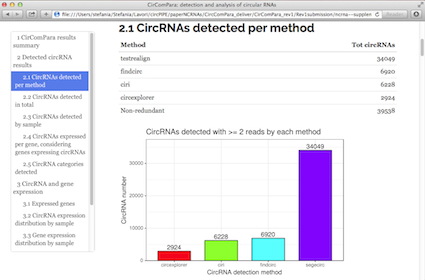
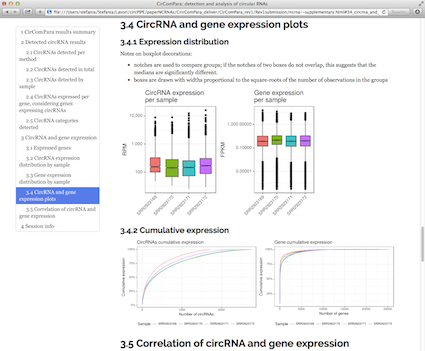
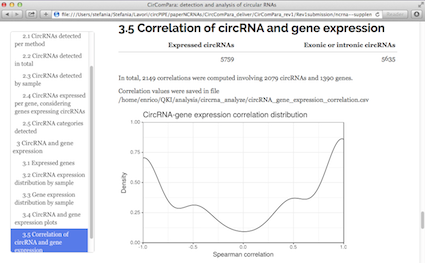
CirComPara is an automated bioinformatics pipeline to detect, quantify and annotate circRNAs from RNA-seq data using in parallel four different methods (CIRCexplorer, CIRI, find_circ, and testrealign) for back-splice identification. CirComPara provides also quantification of linear RNAs and gene expression, ultimately comparing and correlating circRNA and gene/transcripts expression levels.
Citation
Please cite: Gaffo E., Bonizzato A, te Kronnie G., Bortoluzzi S. CirComPara: a multi-method comparative bioinformatics pipeline to detect and study circRNAs from RNA-seq data.
NCRNA Journal, 2017, 3(1), 8; doi:10.3390/ncrna3010008
Special Issue: Bioinformatics Softwares and Databases for Non-Coding RNA Research.