
A-MADMAN: Annotation-based MicroArray DataMeta ANalysis tool
A-MADMAN is an open source web application and gene chip analysis automation framework for annotation-based
 meta-analysis of data from public repositories (NCBI GEO).
meta-analysis of data from public repositories (NCBI GEO).
A-MADMAN allows retrieval, annotation, organization and analysis of gene expression data from public repository.
It automates several analysis steps to be accomplished to setup a working environment for conducting meta-analyses,
without giving up power and flexibility.
A-MADMAN supports a collaborative working style for local or geographically dispersed teams through LAN or Internet
deployment options, but can be used also by a single researcher on his Windows® Personal Computer installing an all-in-one package
that bundles all required dependencies except R. The software, written in python, is based on the popular Django
web framework and uses GNU R as a backend. The package and supporting material can be freely downloaded.
Download A-MADMAN
- Source code (for GNU/Linux systems).
- Windows installer (A self installing package for Windows systems).
Documentation
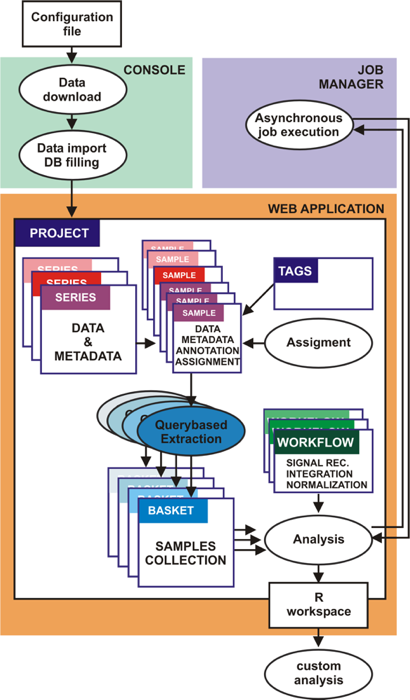
A-MADMAN architecture and flow chart of analysis schema
A-MADMAN include console, job server and web-application. The console allows data retrieval,
import and database filling. The web application, is the user friendly and collaborative core
of the system allowing data inspection, annotation and analysis. A Project is a collection of samples,
series, tags, baskets and analyses owned by a user or by a group of users. Series and samples data and
metadata come from GEO. The user can create an annotation system based on tags and assign samples to
individuals. Queries on Boolean combination of annotation tags are used to select and extract groups
of samples, giving rise to baskets. One or more baskets are passed to a customisable analysis workflow
outputting an R workspace, facilitating following analyses. The job server is in charge for asynchronous
execution of possibly heavy jobs.
