|
- Cancer exome analysis pipeline,
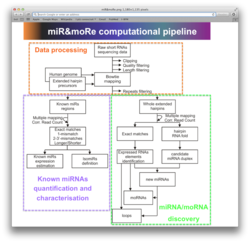
A pipeline for the analysis of exome data for the
identification of relevant somatic variants, mutated
genes and pathways in cancer/normal matched
samples. The pipeline uses gold standard tools,
databases and custom made scripts for calling,
filtering and prioritizing genomic variants from
exome sequencing data (both Illunima and Ion
Proton). Variants are prioritized according to their
recurrence in the samples, predicted effect on
transcripts and annotation in disease databases.
The mutation recurrence of higher order biological
entities, genes and gene network in particular, is
also evaluated to circumvent the classical
scenario in which is very uncommon to identify
variants recurrently mutated in cohorts of patients
harboring the same disease.
prediction of gene silencing effects
- Reference:
Coppe et al. Blood e-letter 2016; Coppe et al. Under evaluation
. 2012
|