SCIENTIFIC ACTIVITY (SECOND YEAR)
The project is carried out in teamwork. During the second year one formal meeting, for progress report, plus several informal meetings involving subgroups of participants, have been organized. Personnel and travel expenses for participation to scientific meetings related to the project (Table 2), cover the major costs of the second year whereas equipment was mainly acquired in the first year, as appropriate.
New collaborations have been set up and new lines of research opened, thanks also to meetings and seminaries organized in relation to the project topics. For instance, a new collaboration of Dr. S. Bortoluzzi with Dr. Te Kronnie (Univ. of Padova) and Prof. L. Meyer (Ulm University, Germany) started an international research project regarding circular RNAs in hematologic malignancies. Moreover, the group leader submitted several applications for International projects specifically on MPN research, in collaboration with both national enterprises and international research partners, including a SME instrument (Horizon 2020, 2014 call) project and a Qatar National Research Fund project.
The website dedicated to the omics4MPN project (compgen.bio.unipd.it/bioinfo/omics4mpn), created to gather and disseminate main results and products of our scientific activity, including publications, software, facilities, has been updated and filled with new data.
Planned activity of the third year
Currently we are completing with analyses, interpretation and experiments the results of the two lines of research roughly corresponding to WP1 and WP2 and the corresponding manuscripts are in preparation. Other methodological papers are under evaluation or in preparation.In the next and last year of the project we plan to correlate validated miRNA in PMF granulocytes and their predicted target genes by quantification of their respective levels as a first step.
Then, we will consider both miRNas identified using the RNA-seq-based approach and those outlined by systems biology analyses of expression data. We will start to address the functional relevance of these selected miRNAs by functional tests using clonogenic assays, analysis of differentiation in liquid cultures and of differentiation-associated markers, analysis of cell cycle and apoptosis.
In parallel, we will continue to work on methodological aspects in the field of both data integration, using a systems biology approach, and RNA-seq analysis issues.
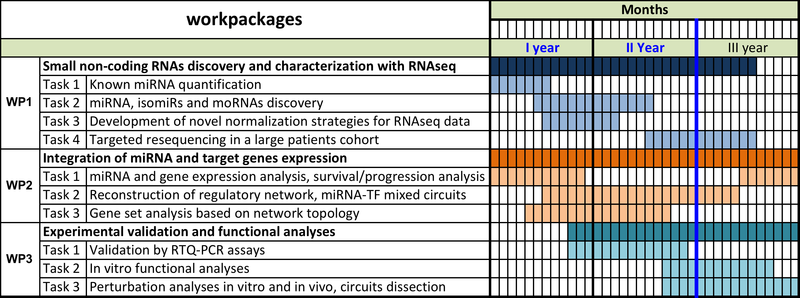
Figure 1. Original project timeline with organization in work packages (WP) and tasks, highlighting the activities planned for the second year.
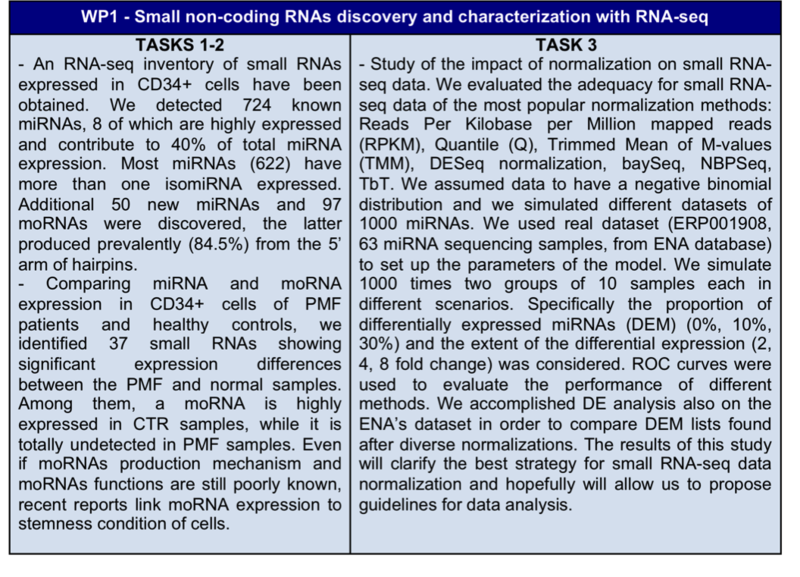

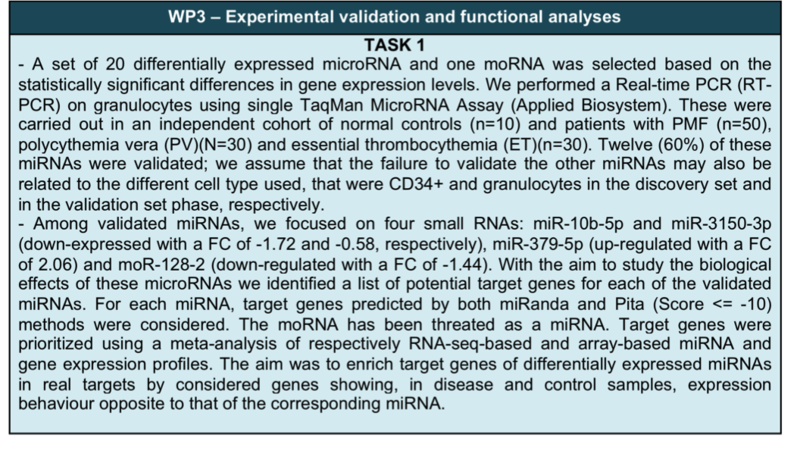
Table 1. Results obtained in the tree planned WPs.
Selected publications
(For an extended list of publications see http://compgen.bio.unipd.it/bioinfo/omics4mpn/dissemination/)- Norfo R, Zini R, Pennucci V, Bianchi E, Salati S, Guglielmelli P, Bogani C, Fanelli T, Mannarelli C, Rosti V, Pietra D, Salmoiraghi S, Bisognin A, Ruberti S, Rontauroli S, Sacchi G, Prudente Z, Barosi G, Cazzola M, Rambaldi A, Bortoluzzi S, Ferrari S, Tagliafico E, Vannucchi AM, Manfredini R; Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators. miRNA-mRNA integrative analysis in primary myelofibrosis CD34+ cells: role of miR-155/JARID2 axis in abnormal megakaryopoiesis. Blood. 2014; 124(13):e21-32. doi: 10.1182/blood-2013-12-544197
- Martini, P., Sales, G., Calura, E., Cagnin, S., Chiogna, M., Romualdi, C. TimeClip: pathway analysis for time course data without replicates. BMC Bioinformatics. 2014; 15(Suppl 5)S3. DOI:10.1186/1471-2105-15-S5-S3
- Calura E, Martini P, Sales G, Beltrame L, Chiorino G, D'Incalci M, Marchini S, Romualdi C. Wiring miRNAs to pathways: a topological approach to integrate miRNA and mRNA expression profiles. Nucleic Acids Res. 2014;42(11):e96. doi:10.1093/nar/gku354.
- Bisognin A, Pizzini S, Perilli L, Esposito G, Mocellin S, Nitti D, ZanovelloP, Bortoluzzi S, Mandruzzato S. An integrative framework identifies alternative splicing events in colorectal cancer development. Mol Oncol. 2014 8(1):129-41. doi: 10.1016/j.molonc.2013.10.004.
- Pennucci V, Zini R, Norfo R, Guglielmelli P, Bianchi E, Salati S, Sacchi G, Prudente Z, Tenedini E, Ruberti S, Paoli C, Fanelli T, Mannarelli C, Tagliafico E, Ferrari S, Vannucchi AM, Manfredini R; on behalf of Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative (AGIMM) Investigators. Abnormal expression patterns of WT1-as, MEG3 and ANRIL long non-coding RNAs in CD34+ cells from patients with primary myelofibrosis and their clinical correlations. Leuk Lymphoma. 2014 Jun 16:1-5.
- Rumi E, Pietra D, Pascutto C, Guglielmelli P, Martínez-Trillos A, Casetti I, Colomer D, Pieri L, Pratcorona M, Rotunno G, Sant'Antonio E, Bellini M, Cavalloni C, Mannarelli C, Milanesi C, Boveri E, Ferretti V, Astori C, Rosti V, Cervantes F, Barosi G, Vannucchi AM, Cazzola M; Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators (including Bortoluzzi S, Bisognin A, Coppe A, and Saccoman C) Clinical effect of driver mutations of JAK2, CALR, or MPL in primary myelofibrosis. Blood. 2014; 124(7):1062-9. doi: 10.1182/blood-2014-05-578435.
- Perilli L, Vicentini C, Agostini M, Pizzini S, Pizzi M, D'Angelo E, Bortoluzzi S, Mandruzzato S, Mammano E, Rugge M, Nitti D, Scarpa A, Fassan M, Zanovello P. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget. 2014; 5(16):6611-9.
- Mussolin L, Holmes AB, Romualdi C, Sales G, D'Amore ES, Ghisi M, Pillon M, Rosolen A, Basso K. An aberrant microRNA signature in childhood T-cell lymphoblastic lymphoma affecting CDKN1B expression, NOTCH1 and growth factor signaling pathways. Leukemia. 2014 Sep;28(9):1909-12. doi: 10.1038/leu.2014.134.
