SCIENTIFIC ACTIVITY (FIRST YEAR)
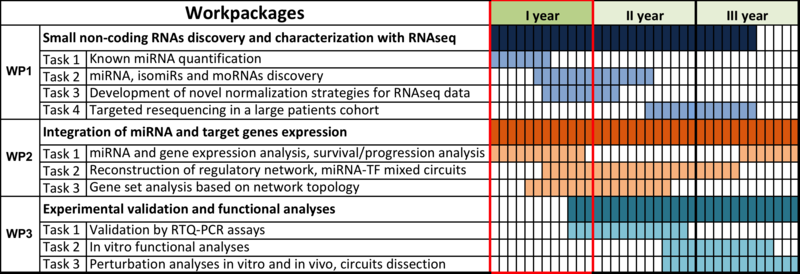
In this report, we present the project results respect to the tasks originally planned for
the first year of activity (Figure 1). All of the planned objectives have been accomplished.
Moreover, additional results have been obtained, as detailed in Table 1 and in Figures 2-3.
Two young researchers specifically recruited and fully dedicated to
the project jointly with additional member of the interdisciplinary team that are working for
the positive development of the project. Three further fellows are expected to start working
in the second year of the project. The project team works in strict collaboration and two formal
meetings for progress report have been organized in Padova during the first year.
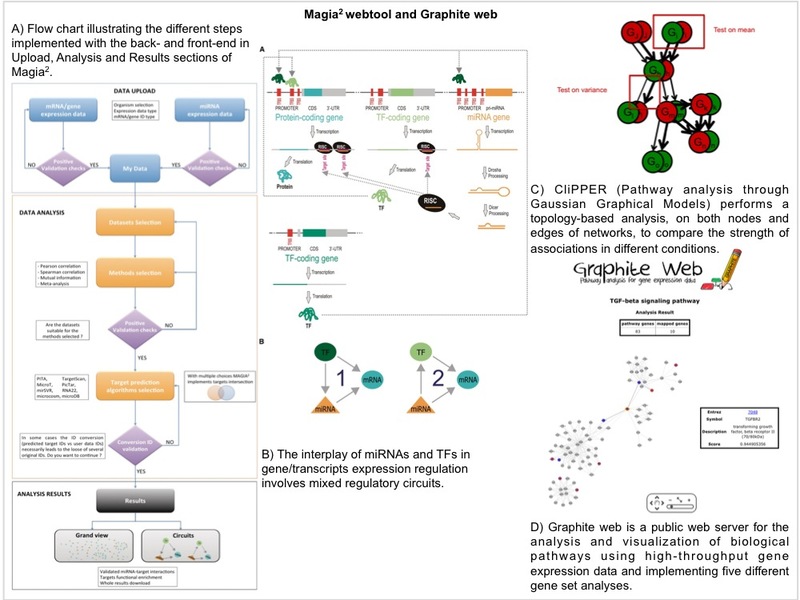
We have integrated our computational infrastructure with new components and we have built
up a new and powerful computational facility that allows storage and analysis of big data sets.
New collaborations have been set up, thanks also to meetings and seminaries organized in relation
to the project topics (Prof. Peter Rogan, Univ. of Western Ontario, Canada; Prof.
Sorin Draghici, Wayne State Univ., Detroit, USA; Prof. Nikolaus Rajewsky,
MDC and Charitè, Berlin and NY Univ., USA). In collaboration with the group Prof.
Vincenzo Bronte (Univ. of Verona) we discovered an unusual regulatory
loop controlling macrophage differentiation, in which the microRNA
miR-142-3p either downregulates canonically gp130 and impacts on the ratio
of the three C/EBPβ transcription factor isoforms, mainly suppressing the activating
LAP∗ isoform; in turn, miR-142-3p transcription is controlled by the isoforms ratio.
The peculiar characteristic of this regulatory circuit is that miR-142-3p exerts its
regulatory activity on C/EBPβ by non canonical binding to the mRNA coding sequence,
in a region including one of three in frame translation initiation sites (aTIS an dTIS1-2)
of the C/EBP mRNA (Sonda et al., Immunity 2013). Starting from this evidence, we are exploring
genome-wide the impact of microRNAs on alternative translation of mRNAs (Bisognin et al. in preparation).
In the remaining part of the project we plan i) to complete the reconstruction of
integrated regulatory networks modulated in PMF for the identification of circuits
altered in the disease, ii) to elucidate by functional assays, carried out
also in a mouse model, the molecular mechanisms involved in disease development.
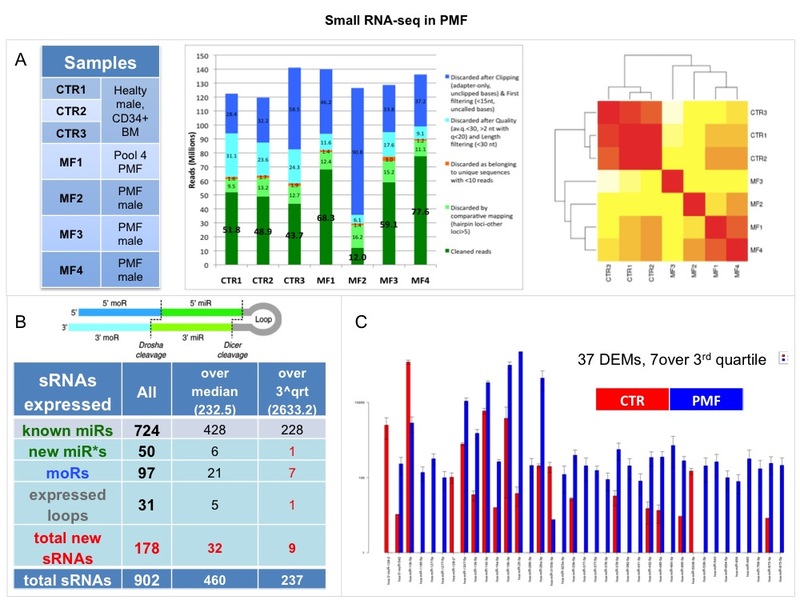
The ongoing validation in about 100 MPN patients of 15 interesting small RNAs and 2
moRNAs using qRT-PCR will be completed and the results will be integrated and interpreted
in conjunction with recent findings stemming from clinical findings (Vannucchi et al, Leukemia 2013).
We will continue to work on several methodological aspects in the field of both data integration,
using a systems biology approach, and RNA-seq analysis issues.